Introduction
Among different uses of the worm model, the strongly standardized survival assays have been proven as very convenient in order to investigate and identify the genetic bases of lifespan as well as the aging processes and related physiological pathways induced by different environmental factors.
Several assays are classically performed: lifespan assays, abiotic stress resistance assays, and pathogen resistance assays. Exact and specific environmental conditions are used to perform survival assays [2-3]. The main aim of those experiments is to understand the factors that regulate complex processes, considering that those processes are strongly conserved among animal evolution, they can provide precious insights into homologous process in more complex animals as well humans.
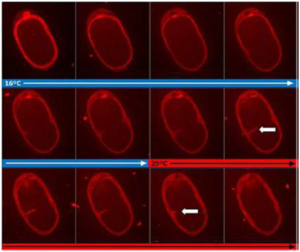
Ultra fast temperature shift device for in vitro experiments under microscopy
Survival assays for lifespan studies
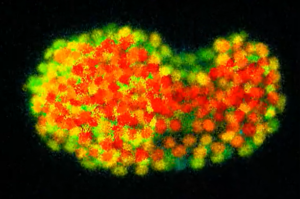
C. elegans survival assays have been key tools for studying physiological processes including: aging, stress resistance and immunity. One noticeable advantage of C. elegans for such assays is that synchronized isogenic populations can be easily obtained since the worm usually exists as a self-fertilizing hermaphrodite that produces hundreds of isogenic progeny (Fig. 2).
Several assays are classically performed: lifespan assays, abiotic stress resistance assays, and pathogen resistance assays. Exact and specific environmental conditions are used to perform survival assays [2-3]. The main aim of those experiments is to understand the factors that regulate complex processes, considering that those processes are strongly conserved among animal evolution, they can provide precious insights into homologous process in more complex animals as well humans.
Worm lifespan assays (or aging assays)
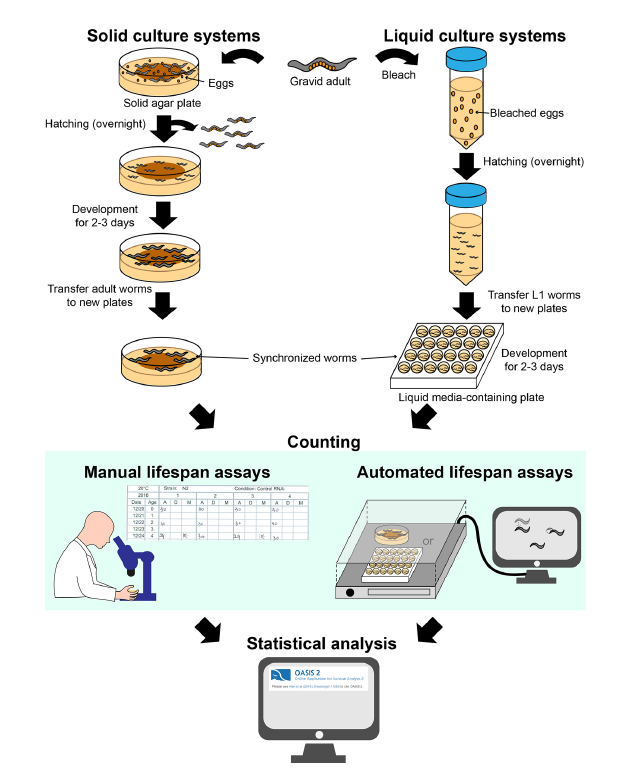
C. elegans is a useful and convenient model for studying aging because of its short lifespan and genetic tractability. Briefly, researchers count live and dead worms in synchronized populations at regular intervals (e.g., 1 or 2 days) and discard dead worms to prevent confusion while performing subsequent counting. Lifespan curves are then generated to illustrate the percentages of live worms in populations over time (Fig 2). There are classically 2 types of systems that are used: solid culture systems and liquid culture systems.
- Solid culture systems
Most commonly used solid culture systems are agar-based hydrogels containing E. coli bacteria as food to support the nematode growth [4]. This approach allows an easy observation of the live/death status of the worm although limitations such as worms that crawl off the plate or burrow inside the agar and the possible desiccation of the plate during the lifespan period can produce bias.
The first step is to synchronize all the worms at the same developmental stage: gravid adult lay eggs that, once hatched, they will originate a cohort of young adults that will be used for the lifespan assay. To discriminate between dead and living specimens, each C. elegans nematode is gently touched by a sterilized platinum wire: dead worm does not react to the stimulus. Alternatively, it is possible to count live worms by their ability to avoid contact with a heated platinum wire [2].
- Liquid culture systems
Media used in liquid culture lifespan assays usually consist of S-basal buffer supplemented with E. coli [4] in order to feed the worms. These types of culture systems are suitable to screen the effect of diet components or chemical treatment on the worms. The main drawbacks of this culturing approach concern the difficulty to handle worms on a platinum wire in a 3D liquid environment and the need for additional equipment (platform, shaker…).
As for all the lifespan experiments, age synchronization at the start of the experiments is a prerequisite. The method adopted to synchronize worms in liquid condition is known as “bleaching” [2] and is based on the capability of eggs to survive to sodium hypochlorite.
Methods for identifying dead worms differ between liquid and solid culture systems. In liquid-based assays, worms that do not display spontaneous movements or respond to shaking are counted as dead. Fluorescent dyes, such as SYTOX, that are able to penetrate cell membranes and emit fluorescent signals only if the specimens are dead can be used to facilitate and eventually automatize the live/dead count [5].
- Automated methods
The previously mentioned approaches have one big drawback. The experiments are manual performed and, besides being time-consuming, they rely on operators skills and experience making them prone to human factor bias. Thus, there is a need for automated methods to overcome these disadvantages.
For solid culture systems, the “WormScan” system can automatically determine the death of worms using image tracking technology [6], whereas the “Lifespan Machine” measure the worm lifespan by processing time-lapse photographs [7].
For the liquid culture systems, microfluidic device called “WormFarm” removes progeny based on differences in worm size and automatically detects death via video recording [8].
Abiotic stress resistance assays
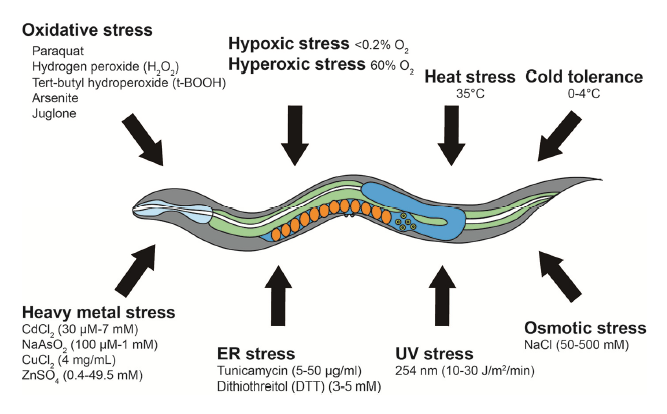
The capability and the responses following a specific stress can provide valuable information regarding the interaction between internal and external stresses as well as the biological processes at the cellular and subcellular level. Stress factors might be induced by oxidative stress, hypoxia, hyperoxia, heat, cold, osmotic, ultraviolet (UV) radiation, endoplasmic reticulum (ER)/unfolded protein reponses, and heavy metal contamination (Fig 3).
- Oxidative Stress
C. elegans oxidative stress resistance assays are performed by treating worms with various chemicals that generate reactive oxygen species (ROS). Due to chemical toxicity, oxidative assays are completed in relatively shorter times than lifespan assays (procedures to run the tests are quite similar to lifespan assyas). Knowing the oxidative species that will be generated is mandatory (superoxide anions, hydrogen peroxide, tert-butyl-hydroperoxide…). The generation of ROS depends on the type of compound that is used (paraquat, arsenite, juglone…). Surprisingly, non-monotonic dose-response curve are often observed following the exposure to ROS stress. While high concentrations reduce the lifespan, low concentrations increase it [9-10]. Thus, choosing appropriate concentrations is essential for testing the effects of ROS-inducing reagents on lifespan and oxidative stress resistance.
- Hypoxic and hyperoxic stresses
Hypoxia and hyperoxia refer to conditions in which specimens are exposed to abnormally low or high levels of oxygen respectively. Hypoxia can be simulated in two distinct approaches: using physical or chemical methods. For physical induction, worms are placed in a sealed hypoxic chamber (O2< 0.2%). In chemical induction, worms are treated with fresh 0.5 M sodium azide, which causes cellular hypoxic responses via inhibiting mitochondrial respiration complex IV. Hyperoxia can be achieved by using a sealed chamber filled with 60% O2. Hypoxic and hyperoxic tests have led to the characterization of key genetic factors, including hypoxia inducible factor 1 (HIF-1), the master regulator of cellular hypoxic responses [11].
- Heat stress
In C. elegans, high temperatures cause cellular disorders, such as neuronal degeneration and heat induced necrotic cell death. C. elegans is equipped with systems that protect cells from the deleterious effects of high temperatures (eg: HSF-1, FOXO, DAF-16…). A typical temperature for acute heat stress resistance (thermotolerance) assays is 35°C (12). Thermotolerance assays require very precise temperature control systems, as uneven temperature distribution in an incubator or a
small temperature change between experiments may cause large variations in survival rates.
- Cold tolerance
C. elegans cold tolerance assays involve synchronized L4 larval stage or young adult worms at specific temperatures ranging from 0°C to 4°C. For survival monitoring, worms exposed to a specific cold temperature for a certain time are transferred to normal culturing temperatures (20°C or 25°C) for 20 min to 1 h for recovery. Dead worms are then counted similarly as described for other survival assays.
- Osmotic Stress
Hyperosmotic shock induces water efflux and protein aggregation, leading to cellular deterioration and disruption of protein homeostasis (proteostasis). Osmotic stress resistance assays are usually performed on agar gels containing various concentrations of NaCl (e.g., 50 mM to 500 mM) using L4 larval or young adult worms at 20°C [13-14].
- UV Stress
Ultraviolet radiation (100 to 400 nm) is known for inducing specific DNA modifications on exposed animals. Liquid media culture systems are not suitable, as liquid absorbs UV thus making difficult to assess the real exposure of the specimens. Ultraviolet light has dose-dependent effects on the health and survival of C. elegans and, in most cases, 10 to 30 J/m2/min of radiation are used [15-16]. Survival against UV radiation can be performed using different stages of worms to determine its stage-specific effects.
- ER unfolded protein stress
Accumulation of unfolded proteins in the ER acts as stress signals that are transmitted to the nucleus. Representative ER stress-inducing agents include tunicamycin (widely used inhibitor of GPT) and dithiothreitol (DTT) (reducing agent that blocks disulfide bond formation). Typical C. elegans ER stress assays are performed over several hours by incubating worms on agar plates supplemented with E. coli in the presence of tunicamycin or DTT [17].
- Heavy metal stress
Bioaccumulation of heavy metals (typically when specific gravity is over 5g/cm3) causes severe diseases in humans, such as poisoning, renal dysfunction and damage to the central nervous system. Molecules that are devolved to reduce heavy metals effects include cysteine-rich metallothionein proteins. Those proteins protect cells by chelating heavy metal ions, they are also strongly conserved among evolution and thus can lead to important insights into the effects of heavy metals accumulation. For C. elegans heavy metal stress assays, specific metal salts are dissolved and mixed with culture media.
Pathogen resistance assays
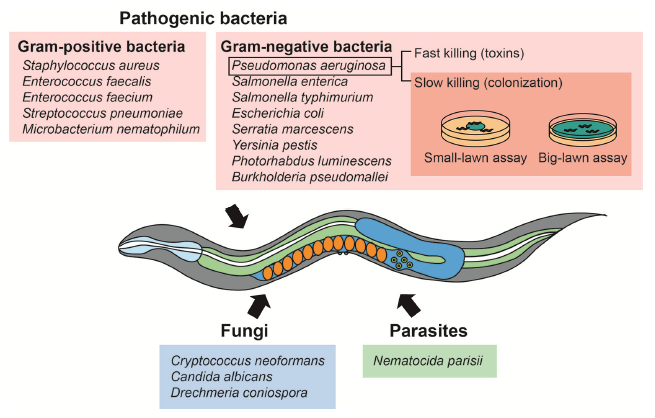
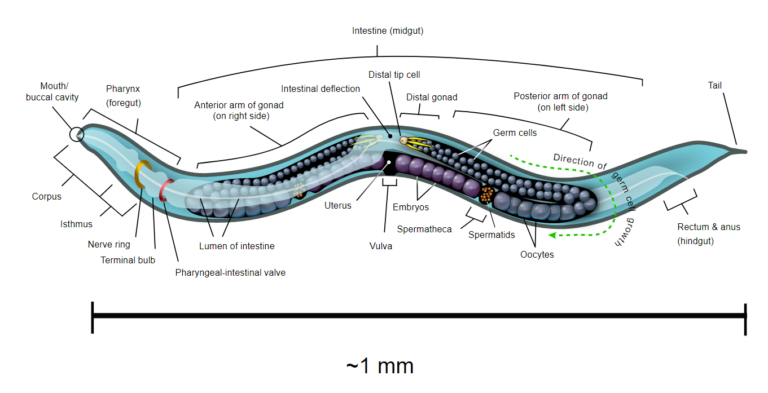
Understanding pathogenesis is also the basis for identifying prophylaxis and treatment strategies against infections. Various organisms, including bacteria, have been used for pathogen resistance assays using C. elegans as a model host (Fig. 4). C. elegans pathogen resistance assays have been also used to investigate the evolution and the basis of pathogenic virulence.
- Bacterial pathogen resistance
Pathogenic bacteria are also mainly divided between Gram-negative and Gram-positive bacteria. Pseudomonas aeruginosa, a ubiquitous Gram-negative bacterium that causes opportunistic infections, is the most popular model pathogen in C. elegans pathogen resistance assays. The methods to conduct a C. elegans survival assays using pathogenic bacteria does not depend on the considered pathogen. P. aeruginosa is the most popular model pathogens in C. elegans pathogen resistance assays. It kills worms through two distinct mechanisms. First, bacterial colonization in the intestine kills C. elegans through quorum sensing and generating multiple virulence factors. Second, P. aeruginosa can rapidly kill C. elegans within 24 h through the secretion of lethal toxins. These two mechanisms have been used for establishing slow- and fast-killing assays. Among the assays described above, slow-killing assays are the most popular survival experiments using P. aeruginosa. Two subtypes of slow-killing assays, the “small-lawn” assay and the “big-lawn” assay, have been used. Performing both small land big-lawn PA14 resistance assays can help identify different mechanisms of pathogen resistance. Several specific considerations for slow-killing should be noted. First, roper cultivation temperatures and media composition, as well as the preparation of fresh bacterial lawns, are crucial for performing reliable assays. Second, using worms in proper developmental stages is critical; young adults and L4 larvae are routinely used for the assays. Third, due to prevalent internal hatching of the worms, FUdR treatment is highly recommended. Fourth, dead worms should be counted every 6 to 12 hours because they quickly lyse and become transparent due to exoenzymes produced by P. aeruginosa.
- Fungi pathogen resistance
Opportunistic fungal pathogens, which are capable of infecting and killing C. elegans, include: Cryptococcus neoformans, Candida albicans, and Drechmeria coniospora. Studies regarding fungal pathogenesis by using C. elegans as the host suggest extensive similarities between fungal virulence factor action in C. elegans and mammals. C. neoformans is a human opportunistic and facultative intracellular fungal pathogen that kills C. elegans by accumulating within its gastrointestinal tract. The fungal culture is spread on plates containing agar gels and incubated overnight. The worms are then transferred onto the fungus-containing plates, and the assays are performed at 25°C.
- Intracellular Parasites
The microsporidian Nematocida parisii is an intracellular pathogen that kills C. elegans. For N. parisii killing assay, the parasite is extracted from the infected worms by vortexing the worms in microcentrifuge tubes containing silicon carbide beads. After filtration using filter paper, the extracts are added to agar plates. Similar to other pathogen resistance assays, L4 larvae or young adults are used for the survival assays at appropriate intervals (e.g., 6 to 12 h) [18].
Conclusion
C. elegans is a very convenient and powerful model to investigate different factors that affect animal lifespan. Survival assay allowed scientists to discover the cellular bases of key aging and stress regulatory factors and to finally and dissect the molecular pathways. Likewise, pathogen resistance assays have shed light on the dynamics implicated in pathogenicity and immunity through the identification of critical virulence factors and immune–regulatory factors. The numerous important discoveries made in the last 40 years and the high standardization of the assays here reviewed make clear that C. elegans will continue to be the reference model leading the way to identify and describe novel regulatory mechanisms crucial for animal physiology.
References
[0] All this review is based on the work of Park HH, Jung Y, Lee SV (2017). Survival assays using Caenorhabditis elegans. Mol Cells 40(2), 90-99
[1] Corsi, A.K., Wightman, B., and Chalfie, M. (2015). A transparent window into biology: a rimer on Caenorhabditis elegans. Worm-Book, 1-31.
[2] Amrit, F.R., Ratnappan, R., Keith, S.A., and Ghazi, A. (2014). The C. elegans lifespan assay toolkit. Methods 68, 465-475.
[3] Keith, S.A., Amrit, F.R., Ratnappan, R., and Ghazi, A. (2014). The C. elegans healthspan and stress-resistance assay toolkit. Methods 68, 476-486.
[4] Stiernagle, T. (2006). Maintenance of C. elegans. WormBook, 1-11
[5] Gill, M.S., Olsen, A., Sampayo, J.N., and Lithgow, G.J. (2003). An automated high-throughput assay for survival of the nematode Caenorhabditis elegans. Free Radic. Biol. Med. 35, 558-565.
[6] Mathew, M.D., Mathew, N.D., and Ebert, P.R. (2012). WormScan: a technique for high-throughput phenotypic analysis of Caenorhabditis elegans. PLoS One 7, e33483.
[7] Stroustrup, N., Ulmschneider, B.E., Nash, Z.M., López-Moyado, I.F., Apfeld, J., and Fontana, W. (2013). The Caenorhabditis elegans lifespan machine. Nat. Methods 10, 665-670.
[8] Xian, B., Shen, J., Chen, W., Sun, N., Qiao, N., Jiang, D., Yu, T., Men, Y., Han, Z., Pang, Y., et al. (2013). WormFarm: a quantitative control and measurement device toward automated Caenorhabditis elegans aging analysis. Aging Cell 12, 398-409.
[9] Hwang, A.B., Ryu, E.A., Artan, M., Chang, H.W., Kabir, M.H., Nam, Survival assays using C. elegans
[10] Lee, S.J., Hwang, A.B., and Kenyon, C. (2010). Inhibition of respiration extends C. elegans life span via reactive oxygen species that increase HIF-1 activity. Curr. Biol. 20, 2131-2136.
[11] Powell-Coffman, J.A. (2010). Hypoxia signaling and resistance in C. elegans. Trends Endocrinol. Metab. 21, 435-440.
Lithgow, G.J., White, T.M., Hinerfeld, D.A., and Johnson, T.E. (1994).Thermotolerance of a long-lived mutant of Caenorhabditis elegans. J. Gerontol. 49, B270-276.
[12] Lamitina, T., Huang, C.G., and Strange, K. (2006). Genome-wide RNAi screening identifies protein damage as a regulator of osmoprotective gene expression. Proc. Natl. Acad. Sci. USA 103, 12173-12178.
[13] Rohlfing, A.K., Miteva, Y., Hannenhalli, S., and Lamitina, T. (2010). Genetic and physiological activation of osmosensitive gene expression mimics transcriptional signatures of pathogen infection in C. elegans. PloS one 5, e9010.
[14] Murakami, S., and Johnson, T.E. (1996). A genetic pathway conferring life extension and resistance to UV stress in Caenorhabditis elegans. Genetics 143, 1207-1218.
[15] Wang, D., Liu, P., and Xing, X. (2010). Pre-treatment with mild UV irradiation increases the resistance of nematode Caenorhabditis elegans to toxicity on locomotion behaviors from metal exposure. Environ Toxicol Pharmacol 29, 213-222.
[16] Shen, X., Ellis, R.E., Lee, K., Liu, C.Y., Yang, K., Solomon, A., Yoshida, H., Morimoto, R., Kurnit, D.M., Mori, K., et al. (2001). Complementary signaling pathways regulate the unfolded protein response and are required for C. elegans development. Cell 107, 893-903.
[17] Troemel, E.R. (2016). Host-microsporidia interactions in Caenorhabditis elegans, a model nematode host. Microbiol Spectr 4.