For the first time Zhuo Du et al. correlate mRNA and protein expression in C. elegans embryo to build the first embryo protein mapping.
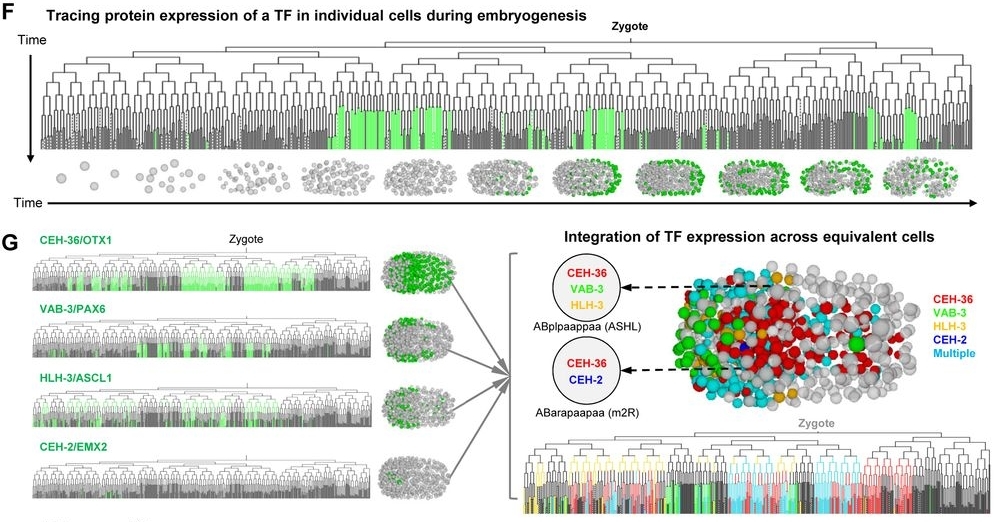
After the generated single-cell mRNA expression atlas of Jonathan S. Packer (2019) in C. elegans embryo, a mapping of proteins expression to better understand biological processes, especially during embryogenesis of C. elegans, has been made by Zhuo Du et al.. A representation encompassing 266 transcription factors allows them to establish a regulatory hierarchy between many lineages, tissues and transcription factors in spatiotemporal fate patterning; also highlighting new functions of certain transcription factors. Since the protein level cannot be directly correlated to the expression of an mRNA.
Check out, how they have tackled this challenge.
Ultra fast temperature shift device for in vitro experiments under microscopy
Abstract
A high-resolution protein atlas is essential for understanding the molecular basis of biological processes. Using protein-fusion reporters and imaging-based single-cell analyses, we present a protein expression atlas of C. elegans embryogenesis encompassing 266 transcription factors (TFs) in nearly all (90%) lineage-resolved cells. Single-cell analysis reveals a combinatorial code and cascade that elucidate the regulatory hierarchy between a large number of lineage-, tissue-, and time-specific TFs in spatiotemporal fate patterning. Guided by expression, we identify essential functions of CEH-43/DLX, a lineage-specific TF, and ELT-1/GATA3, a well-known skin fate specifier, in neuronal specification; and M03D4.4 as a pan-muscle TF in converging muscle differentiation in the body wall and pharynx. Finally, systems-level analysis of TF regulatory state uncovers lineage- and time-specific kinetics of fate progression and widespread detours of the trajectories of cell differentiation. Collectively, our work reveals a single-cell molecular atlas and general principles underlying the spatiotemporal patterning of a metazoan embryo.
References
- Zhuo DU et al., biorxiv.org (2020)