Introduction
Drug induced liver injury (DILI)is one of the major reasons of drug attrition and it mainly occurs because of preclinical trial failure. A possible approach to improve the low predictive power of drug screening could be to find new phenotypic biomarkers of the liver. To do that, super resolution microscopy techniques, also known as nanoscopy, result extremely powerful. In fact, nanoscopy allows detection of structures such as fenestrations (liver nanopores) whose behavior is still unexplained since their size is smaller than the diffraction limit of light. Fenestrations select molecules that are processed by the liver (drugs included) and their role both in physiological and pathological conditions is proved. The nanoscopy techniques developed in the last decade are also compatible with life cells investigation.
How to culture vascularized & immunocompetent 3D models in a standard Multiwell
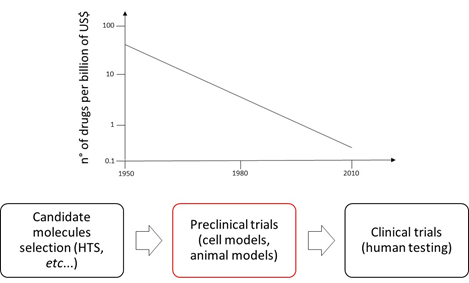
Drug discovery and development scenario
Nowadays pharmaceutical companies spend at least 10 years and 2.5billions of US$ before entering a new drug on the market [1,2]. Albeit the reasons for this situation are many, the major one is the low predictive power of the preclinical test. In fact, 90% of the compounds which pass the preclinical screening fail the human clinical trials, furthermore, the 40% of these failures are due to drug induced liver injury (DILI) [1,3-5]. During preclinical trials drugs are tested on cell and animal models. Cell models generally consist of 2D cell monoculture that, even if cheap and easily manageable, cannot physiology of a complex organ such as liver. Animal testing is not accurate owing to the phylogenetic distance between species which, in several cases, implies different metabolic pathways [1-6,8,9]. This scenario might be mitigated by introducing new systems that better simulate liver physiology (e.g., in vitro liver models and liver on a chip) and combine them with highly effective monitoring apparatus (e.g., super resolution microscopy).
Liver fenestrations
Liver is the largest internal organ of the human body and it has a central role in maintaining homeostasis, metabolic functionality and drug metabolism. Lobules are its fundamental units and they are perfused by capillaries called sinusoids. These microvessels are formed by particular cells, known as liver sinusoidal endothelial cells (LSECs). LSECs have peculiar characteristics that make them considered as a scavenger system and a selective filter for substances passing from the sinusoid lumen to the parenchyma (hepatocytes, fat-cells) and vice versa. In fact, LSECs contain several ultrastructures dedicated to endocytotic activity. Furthermore, they devoid of basal lamina and present nanopores through their surface. These nanopores, also called fenestrations (from Latin, fenestrae), have a 150 to 175 nm diameter and are grouped forming sieve plates which enable the passage of several molecules (e.g., metabolites, plasma proteins, drugs, lipoproteins, viruses, exosomes) from/to sinusoidal lumen to/from hepatocytes. Fenestrations are dynamical structures and their number can change in response to various stimuli and conditions (hormones, drugs, toxins, diseases). LSECs are involved in many liver processes such as chronic liver diseases or liver regeneration. Although the relevance of the alteration in the number of fenestrations is demonstrated for all these processes, many aspects of fenestration’s behavior are still not elucidated. For instance, we do not know how they originated and what determines their structure and size. The understanding of these unresolved aspects would be crucial in order to make out fundamentals of liver biology and elaborate new therapeutic approaches may be based on the sieve’s porosity of the LSECs [10,11,12].
Nanoscopy in liver fenestration studies
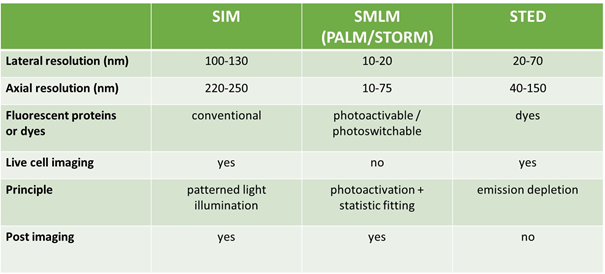
The mean diameter of the liver nanopores through the surface of LSECs ranges from 150 to 175 nm. Such a small size makes fenestrations impossible to be imaged with standard optical microscopy owing to the well-known limitations imposed by the diffraction limit of light. Thus, electronic microscopy (EM) and atomic force microscopy (AFM) was used to study liver fenestrations. Unfortunately, these techniques are technically demanding and not fully compatible with live cells investigation, which is a decisive approach for better comprehending liver nanopore nature. But during the last years other nanoscopy systems were developed. Structured illumination microscopy (SIM), single molecule localization microscopy (SMLM) and stimulated emission depletion (STED) are promising methods that ensure liver nanopores visualization. Furthermore, these allow live cells applications and provide multicolor images. Each technique adopts a different strategy. SIM can detect particles with a size of 100 nm thanks to the phase modulation of visible light obtained through a moving grid with high frequency pitch. SMLM takes advantage of particular fluorophores whose emission is separated in time and space in such a way that they can be distinguished one from the other even though they are very close (20 nm). STED relies on reducing the spot size of the detecting light beam and, in that way, only very close fluorophores are revealed. Nowadays, nanoscopes are primarily used by physicist and bio-physicists but they are expected to be included in most biological laboratories in the next 10 years since they are powerful and suitable for studying live cells [10,15,16].
Conclusion
To date we are not able to predict with high accuracy the drug induced liver injury. The possibility of obtaining new liver biomarkers for drug testing could be a valuable advantage. Liver fenestrations represent a good biomarker candidate, but they are not detectable, especially in living cells. Fortunately, recent nanoscopy techniques (SIM, SMLM, STED) have great potential in revealing liver nanopores. In fact, they can provide multicolor images of fenestrations and can be also used in real time investigations.
References
[1] Khetani, S. R., Berger, D. R., Ballinger, K. R., Davidson, M. D., Lin, C., & Ware, B. R. (2015). Microengineered liver tissues for drug testing. Journal of laboratory automation, 20(3), 216-250.
https://journals.sagepub.com/doi/abs/10.1177/2211068214566939
[2] Zhang, B., & Radisic, M. (2017). Organ-on-a-chip devices advance to market. Lab on a Chip, 17(14), 2395-2420.
https://pubs.rsc.org/ru/content/articlelanding/2017/lc/c6lc01554a/unauth#!divAbstract
[3] Ewart, L., Dehne, E. M., Fabre, K., Gibbs, S., Hickman, J., Hornberg, E., … & Marx, U. (2018). Application of microphysiological systems to enhance safety assessment in drug discovery. Annual review of pharmacology and toxicology, 58, 65-82.
https://www.annualreviews.org/doi/abs/10.1146/annurev-pharmtox-010617-052722
[4] Poloznikov, A., Gazaryan, I., Shkurnikov, M., Nikulin, S., Drapkina, O., Baranova, A., & Tonevitsky, A. (2018). In vitro and in silico liver models: Current trends, challenges and opportunities. ALTEX-Alternatives to animal experimentation, 35(3), 397-412.
https://www.altex.org/index.php/altex/article/view/762
[5] Brown, G. E., & Khetani, S. R. (2018). Microfabrication of liver and heart tissues for drug development. Phil. Trans. R. Soc. B, 373(1750), 20170225.
http://rstb.royalsocietypublishing.org/content/373/1750/20170225
[6] Zheng, F., Fu, F., Cheng, Y., Wang, C., Zhao, Y., & Gu, Z. (2016). Organ‐on‐a‐Chip Systems: Microengineering to Biomimic Living Systems. Small, 12(17), 2253-2282.
https://onlinelibrary.wiley.com/doi/abs/10.1002/smll.201503208
[7] Scannell, J. W., Blanckley, A., Boldon, H., & Warrington, B. (2012). Diagnosing the decline in pharmaceutical R&D efficiency. Nature reviews Drug discovery, 11(3), 191.
https://www.nature.com/articles/nrd3681
[8] Esch, E. W., Bahinski, A., & Huh, D. (2015). Organs-on-chips at the frontiers of drug discovery. Nature reviews Drug discovery, 14(4), 248.
https://www.nature.com/articles/nrd4539
[9] Polini, A., Prodanov, L., Bhise, N. S., Manoharan, V., Dokmeci, M. R., & Khademhosseini, A. (2014). Organs-on-a-chip: a new tool for drug discovery. Expert opinion on drug discovery, 9(4), 335-352.
https://www.tandfonline.com/doi/abs/10.1517/17460441.2014.886562
[10] Øie, C. I., Mönkemöller, V., Hübner, W., Schüttpelz, M., Mao, H., Ahluwalia, B. S., … & McCourt, P. (2018). New ways of looking at very small holes–using optical nanoscopy to visualize liver sinusoidal endothelial cell fenestrations. Nanophotonics, 7(3), 575-596.
https://www.degruyter.com/view/j/nanoph.2018.7.issue-3/nanoph-2017-0055/nanoph-2017- 0055.xml?format=INT&intcmp=trendmd
[11] Braet, Filip, and Eddie Wisse. “Structural and functional aspects of liver sinusoidal endothelial cell fenestrae: a review.” Comparative hepatology 1.1 (2002): 1.
https://comparative-hepatology.biomedcentral.com/articles/10.1186/1476-5926-1-1
[12] Poisson, Johanne, et al. “Liver sinusoidal endothelial cells: physiology and role in liver diseases.” Journal of hepatology 66.1 (2017): 212-227.
https://www.sciencedirect.com/science/article/pii/S0168827816303336
[13] Svistounov, Dmitri, et al. “The relationship between fenestrations, sieve plates and rafts in liver sinusoidal endothelial cells.” PLoS One 7.9 (2012): e46134.
https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0046134
[14] Huang, Bo, Mark Bates, and Xiaowei Zhuang. “Super-resolution fluorescence microscopy.” Annual review of biochemistry 78 (2009): 993-1016.
https://www.annualreviews.org/doi/abs/10.1146/annurev.biochem.77.061906.092014
[15] Erfle, Holger, ed. Super-Resolution Microscopy: Methods and Protocols. Humana Press, 2017.
https://link.springer.com/book/10.1007%2F978-1-4939-7265-4
[16] Yamanaka, Masahito, Nicholas I. Smith, and Katsumasa Fujita. “Introduction to super-resolution microscopy.” Microscopy 63.3 (2014): 177-192.
https://academic.oup.com/jmicro/article/63/3/177/1989177