A fundamental breakthrough in the understanding of development through the realization of the complete transcriptome of C. elegans, from the embryo to the terminal state cell differentiation. Opening the gate for vertebrate development potential disease understanding.
During the development, expression of genes influences cell fate with impacts on the organism later in life. To better understand the fundamental mechanisms of development, gaining new knowledge about gene expression timing and pathways is necessary. Jonathan S. Packer et al. have identified almost all cells terminal and pre-terminal cell types by following the trajectories and temporal dynamics of their respective gene expression. This tracking during C. elegans embryogenesis has allowed these researchers to identify most single-cell transcriptomes. A breakthrough that they hope will lead to new projects and discoveries in vertebrate and potentially human.
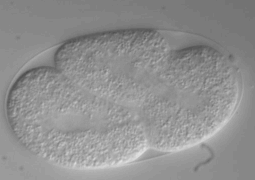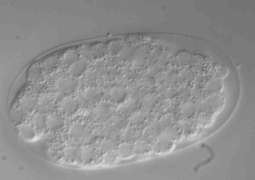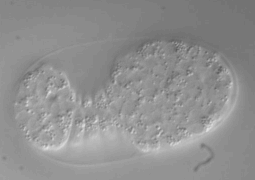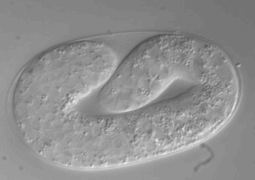
Ultra fast temperature shift device for in vitro experiments under microscopy
Abstract
Caenorhabditis elegans is an animal with few cells but a wide diversity of cell types. In this study, we characterize the molecular basis for their specification by profiling the transcriptomes of 86,024 single embryonic cells. We identify 502 terminal and preterminal cell types, mapping most single-cell transcriptomes to their exact position in C. elegans’ invariant lineage. Using these annotations, we find that (i) the correlation between a cell’s lineage and its transcriptome increases from middle to late gastrulation, then falls substantially as cells in the nervous system and pharynx adopt their terminal fates; (ii) multilineage priming contributes to the differentiation of sister cells at dozens of lineage branches; and (iii) most distinct lineages that produce the same anatomical cell type converge to a homogenous transcriptomic state.