The Human Microbiota present in the body
Trillions of microorganisms habit the human body. Bacteria, fungi, viruses and archaea that are part of the human microbiota colonize the skin, the gastrointestinal tract, the airway tract, the sexual organs, and the eyes. The microbiota plays a crucial role in human body for the synergic functions in metabolic homeostasis, immunology and endocrine functions. in vitro Culturing
On 2007, the National Institute of Health has launched the Human Microbiome Project (HMP) with the aim of study the microbial flora in human in health and disease states, starting from the microbioma (the genoma of the microorganisms). In fact, nowadays, the advances in the next-generation sequencing and in bioinformatic database have revealed the presence of different microorganisms that habit the human body. The HMP has studied the microbioma in skin, urogenital tract, oral mucosa, and nasal mucosa [1]. An emergent interest in ocular surface microbiota and the effect of gut microbiota in eye and brain has lead on new research field and new clinical investigations[2]. in vitro Culturing
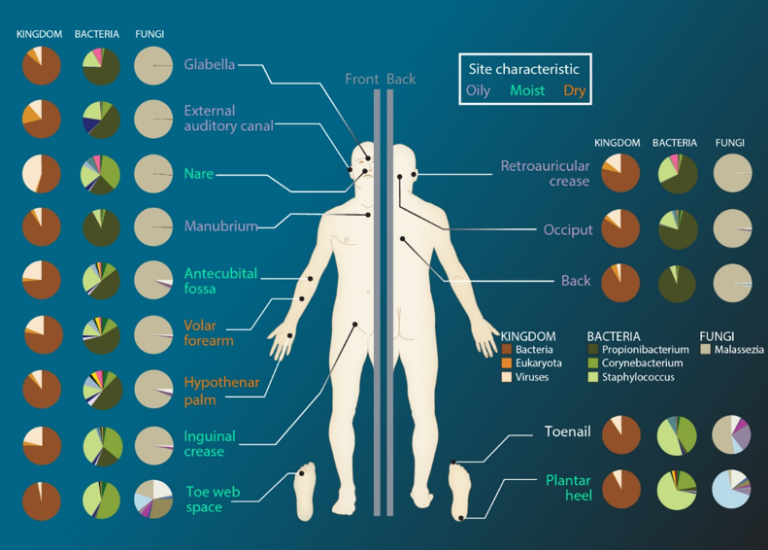
The skin flora or skin microbiota counts commensal or mutualistic bacteria from 19 phyla, counting around of 1000 species[3](Figure 1). The oral, upper digestive, and upper respiratory tract microbioma sequences can be found in the human oral microbioma database and its extension (HOMD and eHOMD). These repositories created by the HMP contains information of about 772 procaryotic species, of which just the 30% is uncultivable and just the 57% of the remaining 70% has been assigned to a known species[4].
How to culture vascularized & immunocompetent 3D models in a standard Multiwell
The Human Microbiota in diseases
Understanding the role of the trillions of bacteria, viruses, and fungi that comprise the human microbiota is critical to understand a variety of human pathophysiologies. Dysbiosis of the skin microbiota had been associated to atopic dermatitis, rosacea, psoriasis, and acne. In the oral mucosa the biofilm produced by the bacteria form the dental plaque which can turn into tartar and lead to gingivitis or periodontal diseases. Vaginal flora alterations are associated with inflammations, bacterial vaginosis, and yeast infections. In the lung diseases, the changes of the commensal bacteria are associated to the progression of chronic obstructive pulmonary disease, asthma, and cystic fibrosis. In the eye, the use of contact lenses and the general exposure to atmospheric agents and pollutions, the alteration of microbiota is associated with corneal infiltrative events, blepharitis, endoophtalmitis, and infection-associated ocular adnexal neoplasm. The gut microbiota has been implicated in several inflammatory and autoimmune diseases, not only in the intestine but also in other organs [5]–[8].
The Human Microbiota cultivation
Historically, microbiology discoveries had been made thanks to the microscope: the first record of a bacterial cell is in 1663 by Antonie Leeuwenhoek. Classification and advances in the microbiology has been focused in instrumental laboratory microscopy, until Robert Koch postulated the importance of culture media in the 1880s generating the shift in cultured and uncultured bacteria. Many different media had been used to improve the possibility to study bacteria in vitro. The food industry demonstrated that the environmental conditions including extreme high and low temperature and pressure are not suitable for living microorganisms. Recently, advances in the technology related to DNA isolation and direct isolation of genomic DNA with next-generation sequencing has led to new discoveries in the microbiology [9]. However, the next steps will focus on finding new methods to cultivate the microbiota based on the optimal temperature and pressure condition in fluidic microchips.
Discover CubiX and our project related to the micrhttps://innovation.cherrybiotech.com/cubix-microphysiological-systemobiota study: GoC-MM.
References
[1] P. J. Turnbaugh, R. E. Ley, M. Hamady, C. M. Fraser-Liggett, R. Knight, and J. I. Gordon, “The Human Microbiome Project,”Nature, vol. 449, no. 7164, pp. 804–810, 2007.
[2] L. J. Lu and J. Liu, “Human microbiota and ophthalmic disease,” Yale J. Biol. Med., vol. 89, no. 3, pp. 325–330, 2016.
[3] E. A. Grice et al., “Topographical and temporal diversity of the human skin microbiome.Grice, E. A., Kong, H. H., Conlan, S., Deming, C. B., Davis, J., Young, A. C., … Segre, J. A. (2009). Topographical and temporal diversity of the human skin microbiome. Science (New York, N,” Science, vol. 324, no. 5931, pp. 1190–2, 2009.
[4] T. Chen, W. H. Yu, J. Izard, O. V. Baranova, A. Lakshmanan, and F. E. Dewhirst, “The Human Oral Microbiome Database: a web accessible resource for investigating oral microbe taxonomic and genomic information.,” Database (Oxford)., vol. 2010, pp. 1–10, 2010.
[5] G. Malaguarnera et al., “Probiotics in the gastrointestinal diseases of the elderly,” J. Nutr. Heal. Aging, vol. 16, no. 4, 2012.
[6] W. H. W. Tang, D. Y. Li, and S. L. Hazen, “Dietary metabolism, the gut microbiome, and heart failure,” Nat. Rev. Cardiol., vol. 16, no. 3, pp. 137–154, 2019.
[7] M. D. P. Willcox, “Characterization of the normal microbiota of the ocular surface,” Exp. Eye Res., vol. 117, pp. 99–105, 2013.
[8] N. Yissachar et al., “An Intestinal Organ Culture System Uncovers a Role for the Nervous System in Microbe-Immune Crosstalk,” Cell, vol. 168, no. 6, pp. 1135-1148.e12, 2017.
[9] J. Handelsman, “Metagenomics: application of genomics to uncultured microorganisms.,” Microbiol. Mol. Biol. Rev., vol. 68, no. 4, pp. 669–85, Dec. 2004.
FAQ
The human body is inhabited by trillions of microorganisms. This collection of organisms is known as the human microbiota. It is composed of bacteria, fungi, viruses, and archaea. These microorganisms colonise various parts of the body. Examples include the skin, the gastrointestinal tract, the airway tract, sexual organs, and the eyes. The microbiota is considered to have an important function in the human body. It is involved in metabolic homeostasis. It also participates in immunology and endocrine functions. A comprehension of this system is necessary for understanding human health.
The Human Microbiome Project (HMP) was started in 2007 by the National Institute of Health. The project’s objective is to study the microbial flora found in humans. This study is applied to both healthy and diseased states. The project began by examining the microbioma, which is the collective genoma of the microorganisms. Progress in next-generation sequencing and bioinformatics databases has revealed the presence of many different microorganisms that inhabit the body. The HMP has specifically studied the microbioma in several areas. These include the skin, the urogenital tract, the oral mucosa, and the nasal mucosa. There is also new research interest in the microbiota of the ocular surface. The effect of the gut microbiota on the eye and brain is also a field of new clinical investigation.
Information on the oral, upper digestive, and upper respiratory tract microbioma can be found in specific databases. These repositories, known as HOMD and eHOMD, were created by the Human Microbiome Project. They contain information on about 772 prokaryotic species. A large portion of these species remains unstudied. It is noted that 30 percent of the species are considered uncultivable. Of the remaining 70 percent that can be grown, only 57 percent have been assigned to a known species. This shows that a substantial part of the oral microbiota has not yet been classified. The skin microbiota is also diverse. It is estimated to include around 1000 species from 19 different phyla.
A comprehension of the trillions of microorganisms in the microbiota is needed to understand many human pathophysiologies. An imbalance, or dysbiosis, of the microbiota has been associated with various conditions. Skin dysbiosis is linked to atopic dermatitis, rosacea, psoriasis, and acne. In the oral mucosa, bacterial biofilm forms dental plaque, which can result in gingivitis or periodontal diseases. Alterations to the vaginal flora are associated with inflammations and infections. Changes in the lung’s commensal bacteria are linked to the progression of asthma and cystic fibrosis. In the eye, microbiota alterations are associated with various conditions, including blepharitis. The gut microbiota is also linked to inflammatory and autoimmune diseases in the intestine and other organs.
Microbiology discoveries were first made using the microscope. A major change occurred in the 1880s when Robert Koch introduced the use of culture media. This generated a division between bacteria that could be cultured and those that were uncultured. Many different media have since been used to improve the ability to study bacteria in a laboratory setting. More recently, new discoveries in microbiology have been made. These are due to technological progress related to DNA isolation, genomic DNA isolation, and next-generation sequencing. Future work will be directed at finding new methods to cultivate the microbiota. These methods will be based on the most suitable temperature and pressure conditions, likely within fluidic microchips.