Abstract
(Ramadan et al) : Improved in vitro models of human organs for predicting drug efficacy, interactions, and disease modeling are crucially needed to minimize the use of animal models, which inevitably display significant differences from the human disease state and metabolism. Inside the body, cells are organized either in direct contact or in close proximity to other cell types in a tightly controlled architecture that regulates tissue function. To emulate this cellular interface in vitro, an advanced cell culture system is required (Cell-Cell Crosstalk in Organ-on-a-Chip). In this paper, we describe a set of compartmentalized silicon-based microfluidic chips that enable co-culturing several types of cells in close proximity with enhanced cell-cell interaction. In vivo-like fluid flow into and/or from each compartment, as well as between adjacent compartments, is maintained by micro-engineered porous barriers. This porous structure provides a tool for mimicking the paracrine exchange between cells in the human body. As a demonstrating example, the microfluidic system was tested by culturing human adipose tissue that is infiltrated with immune cells to study the role of the interplay between the two cells in the context of type 2 diabetes. However, the system provides a platform technology for mimicking the structure and function of single- and multi-organ models, which could significantly narrow the gap between in vivo and in vitro conditions.
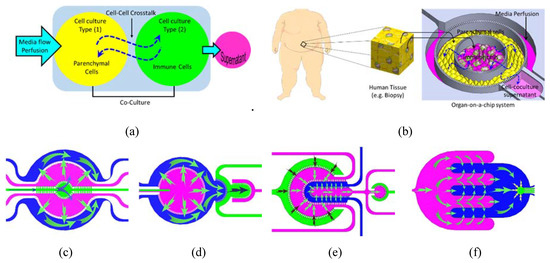
How to culture vascularized & immunocompetent 3D models in a standard Multiwell
Abstract
1. Introduction
Pre-clinical drug screening aims to transform toxicity and efficacy testing from a system reliant on high-dose animal studies to one based primarily on human-relevant in vitro models. The phylogenetic distance between laboratory animals and humans, the discrepancy between current in vitro systems and the human body, and the limitations of in silico modeling have generated the need for new solutions to the ever-increasing demand for safety screening of new substances. The inherent complexity of interconnected tissues in animal models makes it difficult to elucidate and track the physiological events that characterize the interaction between animal organs and exogenic factors. Therefore, improved in vitro models of human organs to predict drug efficacy, interactions, and disease modeling are crucially needed.
2. Cell-Cell Crosstalk in Organ-on-a-Chip
While the simplicity of traditional in vitro models makes them robust and suitable for high throughput research, unfortunately, they provide only little biological relevance to the complex biological tissues of the human body. The validity of an in vitro model is dependent on how it reproduces the key physiological and biological characteristics of the in vivo system. Organs-on-a-chip (OOC) technology has a great potential to mimic many physiological properties of the in vivo organ systems. The ultimate target of the OOC technology is the development of effective and translatable in vitro models with emphasis on investigating physiological events that characterize the interaction between organs, the immune system, and exogenic factors (i.e., pharmaceuticals, chemicals, and nutraceutical stimuli) in health and disease states. Moreover, these systems are powerful tools to provide physiologically relevant in vitro disease models that faithfully reproduce the key physiological aspects of complex human organs.
3. Results and Discussion
3.1. Finite Element Analysis Cell-Cell Crosstalk in Organ-on-a-Chip
The perfusion profile in the compartmentalized chip was simulated with FEA at selected flow configurations by injecting water into selected fluidic inlets and opening selected outlets which correspond to perfusion during the cell incubation time. The FEA meshed models with fine mesh implemented at the porous barriers. The velocity field profile and streamlines show high velocity near inlets/outlets and within the narrow compartments, while the flow velocity profiles tend to be uniformly low within the wide compartments. In general, the velocity observed was at its highest within the sampling compartment of Chip 4 and at its lowest towards the end of the interdigitated fingers of the same chip, while the velocity profile is more uniform across the culture compartments thanks to the perfusion channel arrays. The corresponding shear stresses on the cell membranes were calculated along a central line across the cell culture compartments. The resultant shear stresses in all cell culture compartments were found to be within the range of in vivo physiological interstitial shear stress (≤0.1 dynes per cm2). It should be noted that shear stress can be adjusted by maneuvering the flow rates within the various compartments.
3.2. Microfluidic Testing: Inter-Compartment Permeability
The permeability between the adjacent compartments was investigated using FITC-dextran 4k Da tracer. The tracer was injected into one compartment (upstream) at a concentration of 5 mg/mL until filled with the tracer (observed under the microscope). Then the tracer syringe was replaced by a clear PBS solution and the perfusion was set at a flow rate of 50 µL/min. The fluorescence intensity was recorded at different time intervals across each chip compartment. The recorded intensities and the zone of interest are marked in the inset within the corresponding chip. The FI was measured when the compartments were filled with a clear PBS solution (no FITC-dextran) and subtracted from the total apparent FI to obtain the actual FI due to the dextran permeability. Cell-Cell Crosstalk in Organ-on-a-Chip
3.3. System Testing with Parenchymal-Immune Cell Co-Culture
The OOC chips were tested with an in vitro model that was established and reported in a previous publication, where the experimental procedures are also described. In brief, the biological model comprises a co-culture of human adipocytes (parenchymal) and immune cells which are positioned in two adjutant porous compartments in dynamic culture conditions. Four co-culture experiments were conducted in parallel using the four different chips. Human pre-adipocytes (HPADs) were initially seeded in the designated compartments and the cell growth and morphology were regularly monitored under the microscope. After HPADs reached confluence ~3–5 days after seeding, the cells were perfused with a differentiation medium, and lipid droplets were observed after ~2 days. The differentiated adipocytes are characterized by the presence of different sizes of lipid droplets. Droplet size (hypertrophy) and number (hyperplasia) gradually increased over the 14 days of the differentiation period. To measure the adipocytes hyperplasia, the intracellular content of the cells was stained with fluorescence to enable observing the cell boundary. The lipid droplet number continuously increased during the differentiation period. Hyperplasia was clearly observed during the first week of differentiation of all chips with a significant number of droplets found in Chips 1 and 2. However, during the next two weeks, the individual cell size tended to be increasingly enlarged due to the increase of the lipid droplet size (i.e., the hypertrophy is dominated over the hyperplasia). It shows the highlighted lipid droplets within the adipocytes, which were stained with Oil Red O stain, and it shows the total lipid coverage within the adipocyte compartment which is correlated to the total lipid contents of the adipocytes after 5 and 24 days of adipocyte monoculture. This data confirms the differentiation of preadipocytes to adipocytes (adipogenesis) on-chip. Adipocyte growth and differentiation in the co-culture model were similar to those in the monoculture model. No difference was optically observed.
For immune-metabolic analysis, adipocytes and immune cells were cultured in their designated compartment and the immune-metabolic analysis of the co-culture in Chip 2 was conducted after two weeks of co-culture. By comparing the fluidic and cell co-culture characteristics within the four chips, despite the observed similarity, Chip 1 and Chip 2 showed more uniform fluid flow (Figure 4c,e) and were less prone to bubble generation that could interrupt the cell culture experiments. Due to some chamber narrowing in Chip 2 and Chip 4, the fluid velocity dropped at certain areas, such as within the outer compartment of Chip 3 and at the circular ends of the interdigitated fingers of Chip 4. Therefore, we only present the immune-metabolic results of Chip 2.
The main objective of the current paper is to test various planar arrangements of porous compartments for cell co-culture and to demonstrate a set of designs that can be adapted to selectively position different types of cells in close proximity with efficient intercompartmental crosstalk for organ/disease modeling. Therefore, we only demonstrate the basic functions, i.e., cell co-culture, differentiation, and the basic immune-metabolic profile of the co-culture. In-depth characterization and screening of the biological model were partially reported in a previous publication and are also currently being conducted.
The tested in vitro model mimics the immune cells infiltrate into the human adipose tissue, which is associated with obesity and insulin resistance. It was reported that human adipose tissues express a number of inflammation-related genes with obese subjects displaying a higher expression of these genes as compared to non-obese subjects. These findings support the growing importance of immune-metabolic interplay in human obesity that links to insulin resistance and type 2 diabetes mellitus (T2DM). Cell-Cell Crosstalk in Organ-on-a-Chip
4. Conclusions
A planar compartmentalized microfluidic system with porous sidewalls (barriers) was designed, fabricated, and tested. The microfluidic chip comprises several fluidic compartments which are physically separated by thin sidewalls. The side walls are characterized by a high-density array of small pores with a cross-sectional area smaller than the cell size. The compartmentalized structure enabled the co-culture of two types of cells with each cell type being cultured in its designated compartment. The fluidic compartments are arranged in the planar organization in close proximity to each other, which enables continuous perfusion/diffusion of chemical/biological signals between the cells in the different compartments in a dynamic manner that emulates the environment in the human tissue. The microfluidic system was tested by culturing human adipose tissue that is infiltrated with immune cells to study the role of the interplay between the two cells in type 2 diabetes. However, the system can be employed as a platform for hosting many organ/tissue models.
References
Ramadan Q, Gourikutty SBN, Zhang Q. OOCHIP: Compartmentalized Microfluidic Perfusion System with Porous Barriers for Enhanced Cell–Cell Crosstalk in Organ-on-a-Chip. Micromachines. 2020; 11(6):565. https://doi.org/10.3390/mi11060565