A new microfluidic system allows the high throughput phenotyping of C. elegans embryos!
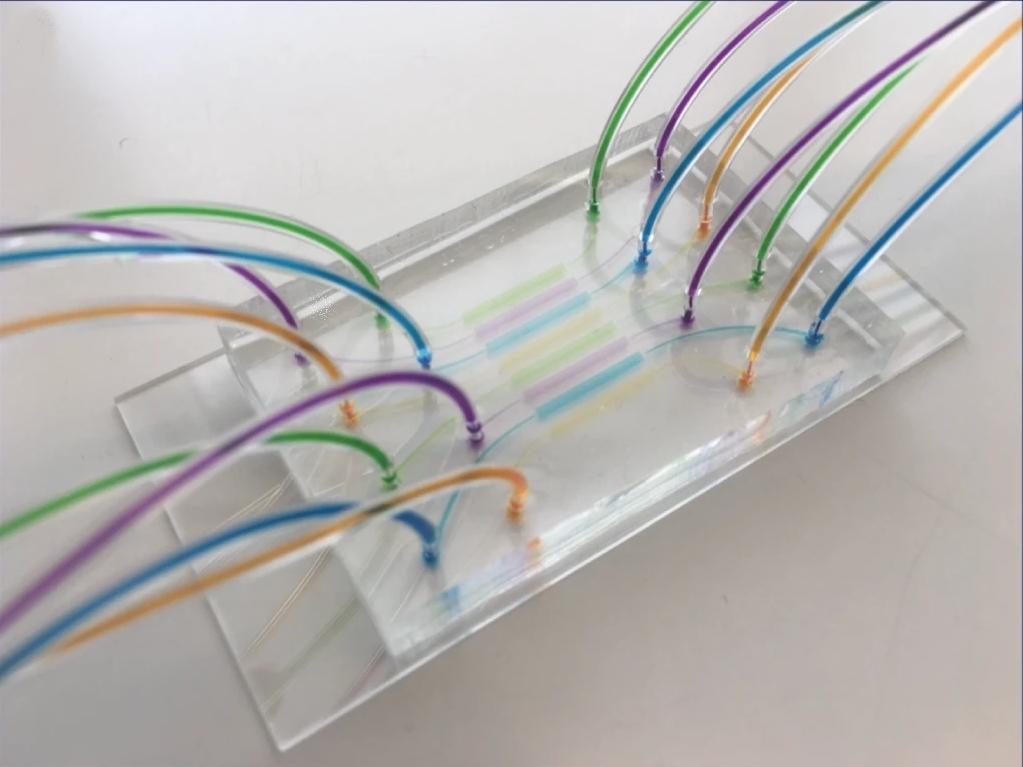
Martin A. M. Gijs et al. produced a high-throughput multiplexed microfluidic platform for automated phenotyping of C. elegans embryos during their full development, in order to make the culture faster and more efficient. Including deep learning and image processing, it produces accurate results in a relatively short amount of time. Each microfluidic lane can create different conditions to simultaneously observe the influence of various compounds on embryonic development.
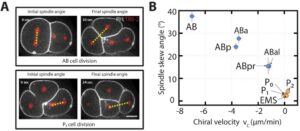
As proof of concept, they made experiences with NaCl and glucose at different concentrations. Results show that the new microfluidic system has been able to analyze developmental lag and lethality due to molarity conditions. After this work, researchers suggest that C. elegans embryos may represent an alternative to adult worms as it leads to comparable experimental conclusions in a shorter time period.
Ultra fast temperature shift device for in vitro experiments under microscopy
Abstract
The nematode Caenorhabditis elegans has been extensively used as a model multicellular organism to study the influence of osmotic stress conditions and the toxicity of chemical compounds on developmental and motility-associated phenotypes. However, the several-day culture of nematodes needed for such studies has caused researchers to explore alternatives. In particular, C. elegans embryos, due to their shorter developmental time and immobile nature, could be exploited for this purpose, although usually their harvesting and handling is tedious.
Here, we present a multiplexed, high-throughput and automated embryo phenotyping microfluidic approach to observe C. elegans embryogenesis after the application of different chemical compounds. After performing experiments with up to 800 embryos per chip and up to 12 h of time-lapsed imaging per embryo, the individual phenotypic developmental data were collected and analyzed through machine learning and image processing approaches. Our proof-of-concept platform indicates developmental lag and the induction of mitochondrial stress in embryos exposed to high doses (200 mM) of glucose and NaCl, while small doses of sucrose and glucose were shown to accelerate development. Overall, our new technique has potential for use in large-scale developmental biology studies and opens new avenues for very rapid high-throughput and high-content screening using C. elegans embryos.
References
- Automated phenotyping of Caenorhabditis elegans embryos with a high-throughput-screening microfluidic platform (Martin A. M. Gijs et al., 2020)